An ORNL–University of Tennessee Graduate School of Medicine collaboration has for the first time successfully characterized the earliest structural formation of the disease type of the protein “huntingtin” that creates such havoc in Huntington’s Disease (HD). The incurable, hereditary neurological disorder is always fatal and affects 1 in 10,000 Americans.
HD is caused by a renegade protein that destroys neurons in areas of the brain concerned with the emotions, intellect, and movement. We all have the normal huntingtin protein, which is known to be essential to human life, although its true biological functions remain unclear. Normal huntingtin contains a region of 10 to 20 glutamine amino acids in succession. However, the DNA of HD patients encodes for 37 or more glutamines, causing instability in huntingtin fragments that contain this abnormally long glutamine repeat. In consequence, the mutant protein fragment cannot be degraded in a normal way and instead aggregates on itself to form deposits of fibrils in neurons.
Researchers originally thought the deposits, that appear under the microscope as “clumps,” were the cause of the devastation that ensues in the brain. More recently they think the clumping may actually be a kind of biological housecleaning, an attempt by the brain cells to clean out these toxic proteins from places where they are destructive. So what are the toxic species? When and where do they occur? To answer this question, Christopher Stanley, a Shull Fellow in the Neutron Sciences Directorate ORNL, began to work with Valerie Berthelier, a UT Graduate School of Medicine researcher who studies protein folding and misfolding in HD. The pair wanted to explore the structures of the earliest aggregate species believed to be the most toxic.
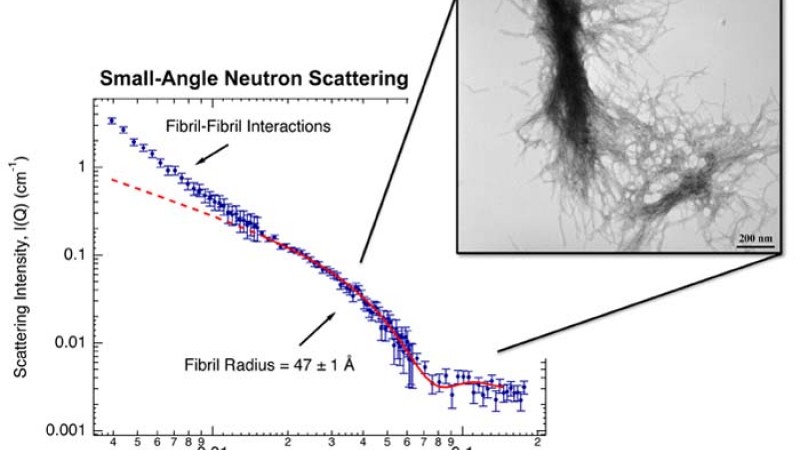
Using the Bio-SANS (small-angle neutron scattering) instrument at ORNL’s High Flux Isotope Reactor (HFIR), Stanley and Berthelier took snapshots of a 42-glutamine mutant huntingtin peptide at Angstrom resolution (tenths of billionths of a meter), as it self-aggregated over time. They were able to determine the size and mass of the mutant protein structures―from the earliest small, spherical precursor species composed of two (dimers) and three (trimers) peptides―along the aggregation pathway to the development of the resulting later-stage fibrils. They were also able to see inside the later-stage fibrils and determine their internal structure, which provides additional insight into how the peptides aggregate.
“This is a great instrument for taking time-resolved snapshots. You can look at how this stuff changes as a function of time and be able to catch the structures at the earliest of times,” Stanley explains. “When you study several of these types of systems with different glutamines or different conditions, you begin to learn more and more about the nature of these aggregates and how they begin forming.”
The samples are aqueous solutions of the peptide contained in cuvettes that have quartz windows. The researchers followed a very careful protocol to ensure that they started with a peptide solution absent of any preformed aggregates. In the current work, they investigated the pathological “disease-relevant” 42-glutamine peptide chain. Their follow-up work involves the study of a normal length 22-glutamine peptide.
At the HFIR Bio-SANS instrument, the neutron beam comes through a series of mirrors that focus it on the sample. The neutrons interact with the sample, probing its atomic structure, and then the neutrons scatter, to be picked up by a detector. From the data the detector sends of the scattering pattern, researchers can deduce the size and shape of the diseased, aggregating protein, at each time step along its growth pathway.
SANS was able to distinguish the small peptide aggregates in the sample solution from the rapidly forming and growing larger aggregates that are simultaneously present. In separate experiments, they were able to monitor the disappearance of the single peptides, as well as the formation of the mature fibrils. SANS also demonstrated the internal composition of the later-stage mature fibrils: loosely packed and quantitatively consistent with the hollow tube-like b-helix model proposed by Nobel Laureate Max Perutz.
“The structural parameters we defined for the early-stage peptides, the dynamics of fibril formation, and the end-state fibrils will be of great value in more carefully discerning their roles in the aggregation pathway,” Stanley says. The research also gives them a path for the development of drugs to treat the condition.
Now that they know the structures, the hope is to develop drugs that can counteract the toxic properties in the early stages, or dissuade them from taking the path to toxicity. “The next step would be, let’s take drug molecules and see how they can interact and affect these structures.”
At the UT Graduate School of Medicine, Berthelier can do in-cell studies to see if cells treated with such drugs live longer or die sooner. At the HFIR Bio-SANS, experiments with drug molecules could discern in what way a drug affects the mutant huntingtin structure. “Is it preventing certain structures from forming, how does it affect aggregation, or is it aggregating at all now? Then we can see in a rational way how it is affecting the protein aggregates, perhaps preventing what can become the disease inside the brain cell,” Stanley says.
For now, the researchers are excited that they have a method that is useful in the further study of HD aggregates and applicable for the study of other protein aggregation processes, such as those involved in Alzheimer’s and Parkinson’s diseases.
“That is the future hope. Right now, we feel like we are making a positive contribution towards that goal,” says the young researcher.
Research funding was provided by the U.S. Department of Energy Office of Biological and Environmental Research; the Clifford Shull Fellowship Program, ORNL (C. Stanley); and NIH grant 1R21NS056325-01A1 (V. Berthelier).
Research was conducted at the High Flux Isotope Reactor, which is funded by the U.S. Department of Energy Office of Basic Energy Sciences.
Reference
:
Christopher B. Stanley, Tatiana Perevozchikova, and Valerie Berthelier, “Structural Formation of Huntingtin Exon 1 Aggregates Probed by Small-Angle Neutron Scattering,” Biophysical Journal 100, 10, May 2011.